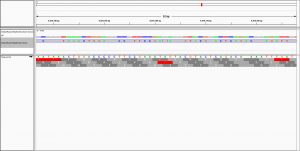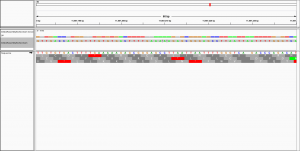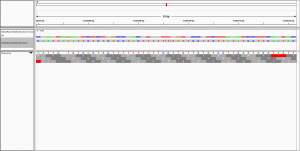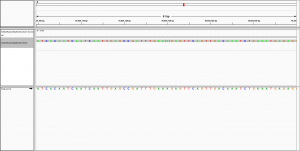
Dear all,
I have indexed the C. elegans reference genome with:
bwa index output/genome/ref/seq/celegans.fa
and then aligned my de novo assembly to the reference with:
bwa mem -t 8 -x intractg output/genome/ref/seq/celegans.fa input/assembly/celegans/hgap/bristol/assembly.fa > output/alignment/pacbio/bwa/ref/bristolAssembly.sam
I then
samtools view -bS output/alignment/pacbio/bwa/ref/bristolAssembly.sam > output/alignment/pacbio/bwa/ref/bristolAssembly.bam
samtools sort output/alignment/pacbio/bwa/ref/bristolAssembly.bam -o output/alignment/pacbio/bwa/ref/bristolAssemblySorted.bam
samtools index output/alignment/pacbio/bwa/ref/bristolAssemblySorted.bam
to visualise the alignment in IGV.